Alternative workflow for COVID-19 diagnosis using direct RT-PCR screening
Fluxo de trabalho alternativo para diagnóstico da COVID-19 utilizando triagem por RT-PCR direta
Wallace Felipe Blohem Pessoa1
Bruno Henrique Andrade Galvão1
Haline Barroso2
Eduardo Sergio Soares Sousa1
Eloiza Helena Campana1
Marília Gabriela dos Santos Cavalvante1
Naiara Naiana Dejani1
Vinícius Pietta Perez1
1Doutor. Universidade Federal da Paraíba (UFPB). João Pessoa, PB, Brasil.
2Mestre. Laboratório Central de Saúde Pública da Paraíba (UFPB). João Pessoa, PB, Brasil.
Suporte Financeiro: The study received financial support from Fundação de Amparo à Pesquisa do Estado da Paraíba (003/2020-FAPESQ/SEECT)
Acknowledgements: To Laboratório Central de Saúde Pública do Estado da Paraíba – LACEN/PB, particularly to Dalane Loudal for assistance.
Recebido em 10/03/2021
Aprovado em 29/04/2021
DOI: 10.21877/2448-3877.202102128
INTRODUÇÃO
Since the first report of SARS-CoV-2, the public health laboratories have worked to develop and validate molecular assays to detect the causative agent of COVID-19.(1) Considered the most effective test for diagnosis in symptomatic individuals, reverse transcriptase PCR (RT-PCR) can help to track positive cases and guide health agents to evaluate potential epidemiological situations.(2) Considering that the vaccines are not yet available for the majority of the population and the lack of effective treatments, detecting the virus and isolating the infected people is the principal tool available to reduce transmission.(3-4) The worldwide COVID-19 infection has passed 105,764,730 cases, Brazil is the third country with highest number of accumulated cases (9,497,795) and the second highest cumulative number of deaths,(5) and has limited access to molecular diagnosis. This is due to few resources and lacking health care policies. Indeed, the pandemic presents diagnostic-supply shortages, notably in developing countries. Thus, we aimed to evaluate and validate the performance of direct RT-PCR (dRT-PCR) as an alternative method to screening COVID-19 during the pandemic.
Material and Methods
Our study included fifty samples (nasopharyngeal swabs) randomly selected from symptomatic individuals at the LaBiMol/CCM/UFPB Laboratory. These were stored in phosphate buffered saline (PBS) at -70°C for COVID-19 testing. According to the BioGene viral DNA/RNA kit (Quibasa, Brazil) manufacturer’s instructions the RNA was extracted using a final elution of 60 μL of DNase/RNase free water. For direct dRT-PCR each sample was inactivated at 95°C for 10 minutes, placed in an ice-bath, and immediately conducted to RT-PCR. Amplification for the SARS-CoV-2 E gene, and human RNAse P was performed according to instructions, using the SARS-CoV-2 kit (E) (Bio-Manguinhos, Brazil). For dRT-PCR, 2 μL of inactivated samples were used, and DMSO added at 2% to complete the final concentration. Reactions were amplified using the QuantStudio3 Real Time PCR System (Thermo Fisher Scientific, USA), and the virus was considered: detectable when the cycle threshold (Ct) < 40 for the E gene, undetectable when Ct > 40 for the E gene and Ct < 35 for RNAse P, and undetermined when Ct > 40 for the E gene and Ct > 35 for RNase P. Results were analyzed statistically, where cycle thresholds were compared using T-test, and concordance in percentage was calculated, Kappa coefficient was used to measure the rate of agreement. Values of p ≤ 0.05 were considered statistically significant. All analyses were performed using IBM SPSS version 20.0 for macOS (IBM Corporation, USA).
Ethics
This research was approved by the Ethics Committee (30658920.4.0000.0008).
Results
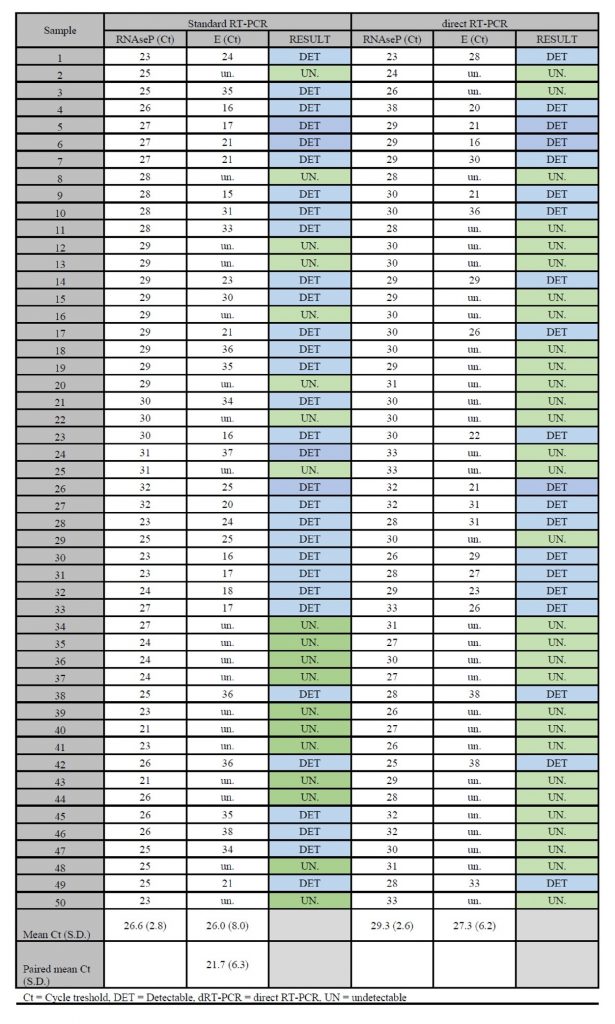
Standard RT-PCR detected the presence of SARS-CoV-2 in 62% of samples (31/50) dRT-PCR detected the virus in 40% (20/50) of these same samples. For both methodologies, all reactions were considered valid and no reactions were undetermined. Concordance between methodologies was 78% (39/50) with moderate agreement (Kappa 0.580 ±0.101, P = 0.001). For dRT-PCR, the positive agreement was 64.5% and negative agreement was 100%. The average Ct detected by dRT-PCR was higher than the average Ct detected by RT-PCR. For RNAse P the values were 26.6 ±2.8 versus 29.3 ±2.6, and for the E gene the values were 21.7 ±6.3 versus 27.3 ±6.2. When comparing the average Ct for the E gene using standard RT-PCR, agreement for the samples observed was at 21.7 ±6.3, disagreement was at versus 33.8 ±3.6 (Chart 1).
Using a serial dilution of clinical samples, the lowest limit detection was established. The relative limit of detection (RLOD) for standard RT-PCR was 10-5 and for direct RT-PCR was 10-4 (Table 1).
Table 1 – Relative Limit of Detection (RLOD) for standard RT-PCR and direct RT-PCR methods for E gene.
| Standard RT-PCR | RLOD | direct RT-PCR | RLOD | |||||||
| Concentration | 102 | 103 | 104 | 105 | 105 | 102 | 103 | 104 | 105 | 104 |
| Pos./total | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 1/5 | ||
| Mean Ct | 23.52 | 26.62 | 31.14 | 34.62 | 28.16 | 32.34 | 35.44 | 37.1 | ||
| Std. Deviation | 0.89 | 1.48 | 0.28 | 0.43 | 0.89 | 0.46 | 0.71 | |||
*Concentration is presented in a serial dilution of clinical samples. The observed Relative Limit of Detection (RLOD) is the lowest concentration where all replicates detect E gene.
Chart 1. Comparative results of standard RT-PCR versus direct RT-PCR for 50 samples included in this study
Discussion
The primer-probe set for the E gene described by Corman et al.(1) is one of the most widely used in Brazilian public health laboratories, with a limit of detection estimated at 3.9 copies per reaction. Several different primer-probe sets are described for detection of SARS-CoV-2, and all have been found to be highly specific with no cross reactions in samples of patients infected with other respiratory viruses. However, the calculated sensitivity is different between primer-probe sets, and assays using the same E gene set evaluated in our study displayed increased sensitivity, thus they are able to reliable detect the viral genome in samples containing 6.3 viral copies and an average Ct of 37.2 (±1.34).(6)
Purification of viral RNA, manually by spin-column, with subsequent amplification using SARS-CoV-2 kit (E) is a time-consuming process. Due to eventual shortage of RNA purification kits, we evaluated a quick alternative dRT-PCR which resulted in perfect negative concordance, and a moderate positive concordance when using the standard procedure. In Denmark, a study with a similar methodology observed sensitivities from 89.5% to 92%, with specificity at 100%, with a mean Ct increment of from +1.4 to +1.9 for dRT-PCR using both the same primer-probe set and the sample inactivation procedure described in our study.(7)
For sample collection, PBS is a viable alternative for transport,(8) but the sensitivity of dRT-PCR might be more affected than standard RT-PCR by the medium used. In Canada, it has been reported that samples stored in a balanced salt solution medium presented a positive agreement of 69% and increased inhibition of the PCR reaction. Employing our methodology of samples stored in PBS, we observed a similar positive agreement of 63.2%, but unlike the results of Merindol et al.(9) no increase in reactions, which did not amplify the internal control. Another study, conducted in Spain, achieved similar results, and all samples amplified the internal control.(10) A plausible explanation is the reduced volume used in ours and the Spanish study, which diluted the PCR inhibitors present in the raw samples.
Due to dilution or to the modified transport media our alternative dRT-PCR resulted in an increased Ct for the E gene (+4.2) and discrepant samples between standard RT-PCR and dRT-PCR presented high average Ct using standard RT-PCR (an average of 35.2). Thus, we may assume that the false negatives encountered using dRT-PCR were related to low viral load.
Other fast and inexpensive alternatives to RNA extraction from nasopharyngeal swabs have also been evaluated. In Hong Kong, samples stored in viral transport medium were heat inactivated, whether preceded or not, by treatment with proteinase K. When pre-treated, an increase in the dRT-PCR detection rate was observed.(11) We also evaluated the use of proteinase K; however, the rate of undetermined reactions was excessively high (data not shown), which suggests that dRT-PCR performance can be affected by many factors.
When comparing the available studies, one may observe that under different protocols, results are not reproduced, and not all RT-PCR kits are compatible with simplified sample heat-processing. The use of dRT-PCR should only be chosen upon proper validation.
The lack of extraction reduces turnaround time for COVID-19 diagnosis, thus allowing prompt decision making regarding isolation of infected patients. However due to the inferior sensitivity of dRT-PCR, it is proposed that dRT-PCR may be used for screening. To optimize time, and consumption of reagents, screening could be performed for the E gene alone, without an internal control (RNAse P); and samples presenting Ct ≥ 40 be submitted for RNA extraction/purification and standard RT-PCR.
To evaluate the technical convenience of the proposed method, total time per run and sample quantity, according to the prevalence observed in our laboratory, were calculated. Using the proposed method resulted in decreased time to detect the virus, yielding results in 2 hours and 20 minutes for 23.3% to 44.4% of samples, and in 8 hours and 20 minutes for 55.6% to 76.7% of samples.
Conclusion
In a sample population with high prevalence, we have demonstrated that dRT-PCR may be used for screening in molecular diagnosis of COVID-19, resulting in reduced RNA viral extraction kit consumption and shortened execution times. The proposed workflow represents a viable alternative towards avoiding RNA purification kit shortages in public health laboratories.
Resumo
Objetivo: A COVID-19 é atualmente um sério problema de saúde pública e o diagnóstico é a principal ferramenta para controlar e monitorar a propagação da doença. Este estudo teve como objetivo avaliar a eficiência da RT-PCR direta (dRT-PCR) para detecção do SARS-CoV-2. Métodos: Vinte e sete amostras de swab nasofaríngeo de indivíduos sintomáticos foram avaliados. A RT-PCR padrão foi realizada e para a dRT-PCR as amostras foram pré-aquecidas antes da amplificação. Resultados: A concordância positiva foi de 63,2% e a concordância negativa foi de 100%, sendo moderadamente concordante. Conclusão: A dRT-PCR pode ser uma alternativa para a triagem de pacientes sintomáticos e uma opção confiável durante uma eventual escassez de kits de purificação de RNA viral.
Palavras-chave:
Virologia; Triagem; Técnicas de Diagnóstico Molecular; Reação em Cadeia da Polimerase
REFERENCES
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045.
- Nörz D, Fischer N, Schultze A, Kluge S, Mayer-Runge U, Aepfelbacher M, Pfefferle S, Lütgehetmann M. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J Clin Virol. 2020 Jul;128:104390. doi: 10.1016/j.jcv.2020.104390.
- Zhang LP, Wang M, Wang Y, Zhu J, Zhang N. Focus on the 2019 novel coronavirus (SARS-CoV-2). Future Microbiol. 2020 Jul;15:905-918. doi: 10.2217/fmb-2020-0063.
- WHO. World Health Organization. Considerations for quarantine of individuals in the context of containment for coronavirus disease (COVID-19). Switzerland: World Health Organization [updated 2020 March 19; cited 2020 July 27]. Available from: https://www.who.int/publications/i/item/considerations-for-quarantine-ofindividuals-in-the-context-of-containment-for-coronavirus-disease-(covid-19
- Ministério da Saúde. Boletim Epidemiológico Especial. Semana Epidemiológica 5 (31/1 a 6/2/2021) [internet]. Brasília: Ministério da Saúde. Secretaria de Vigilância em Saúde. Doença pelo Coronavírus COVID-19. [acesso em março 2021] Disponível em: https://www.gov.br/saude/ptbr/media/pdf/2021/fevereiro/13/boletim_epidemiologico_covid_49_13fev21.pdf
- Nalla AK, Casto AM, Huang MW, Perchetti GA, Sampoleo R, Shrestha L, Wei Y, Zhu H, Jerome KR, Greninger AL. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J Clin Microbiol. 2020 May 26;58(6):e00557-20. doi: 10.1128/JCM.00557-20.
- Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS-CoV-2 – escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. doi: 10.2807/1560-7917.
- Rodino KG, Espy MJ, Buckwalter SP, Walchak RC, Germer JJ, Fernholz E, Boerger A, Schuetz AN, Yao JD, Binnicker MJ. Evaluation of Saline, Phosphate-Buffered Saline, and Minimum Essential Medium as Potential Alternatives to Viral Transport Media for SARS-CoV-2 Testing. J Clin Microbiol. 2020 May 26;58(6):e00590-20. doi: 10.1128/JCM.00590-20.
- Merindol N, Pépin G, Marchand C, Rheault M, Peterson C, Poirier A, Houle C, Germain H, Danylo A. SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J Clin Virol. 2020 Jul;128:104423. doi: 10.1016/j.jcv.2020.104423.
- Alcoba-Florez J, González-Montelongo R, Íñigo-Campos A, de Artola DG, Gil-Campesino H, The Microbiology Technical Support Team, Ciuffreda L, Valenzuela-Fernández A, Flores C. Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int J Infect Dis. 2020 Aug;97:66-68. doi: 10.1016/j.ijid.2020.05.099.
- Chu AW, Chan WM, Ip JD, Yip CC, Chan JF, Yuen KY, To KK. Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits. J Clin Virol. 2020 Aug;129:104519. doi: 10.1016/j.jcv.2020.104519.
Correspondência
Wallace Felipe Blohem Pessoa
Centro de Ciências da Saúde,
Departamento de Fisiologia e Patologia,
Campos I, Cidade Universitária,
Universidade Federal da Paraiba-UFPB
E-mail: [email protected]
